Genomic Diversity:
What is a Genomic Diversity Score: Genomic Diversity Scores can now be calculated for all Japanese Black Fullblood animals registered with the AWA. This is achieved by computing a Genomic Relationship Matrix (GRM), which calculates pairwise comparisons of the individual SNP similarities between all animals in the genomic database. This allows us to understand how each animal is genomically related to every other animal in the database. Animals which are closely related to the population have low Genomic Diversity Scores.
Calculating Genomic Diversity: Firstly, you need a genomic SNP profile. It is important to note that a Genomic Diversity Score can only be calculated from genomic profiles (not MiP, STR or parentage SNP) and therefore, not all animals will get a Diversity Score calculated.
The Diversity Score is calculated by analysing the SNP by SNP similarity in an individual to the reference population. The reference population is set as the last five calendar years of registered Fullblood Japanese Black females, which will be approximately 70% of the future breeding population.
The Diversity Score gets calculated in three steps:
1. the average of the animal’s relationship to all animals in the reference population gets calculated; then
2. this value gets inversed to result in higher values indicating more diversity (low genomic relationship to the reference population); and
3. the value is expressed between 0 and 100, with 0 being lowest diversity and 100 being highest diversity.
Genomic Diversity Scores can currently only be calculated for Fullblood Japanese Black animals that have a genomic profile. We do not have appropriate reference populations for Red Wagyu, Purebred or content populations.
Examples: Genomic Diversity Scores can assist breeders to identify animals that are relatively unrelated to the reference population. This could be used to select unrelated sires to a female herd, to reduce the inbreeding levels in offspring. The following figure shows the distribution of Diversity Scores in all Japanese Black Fullblood registered sires compared to females registered in the last 5 years.
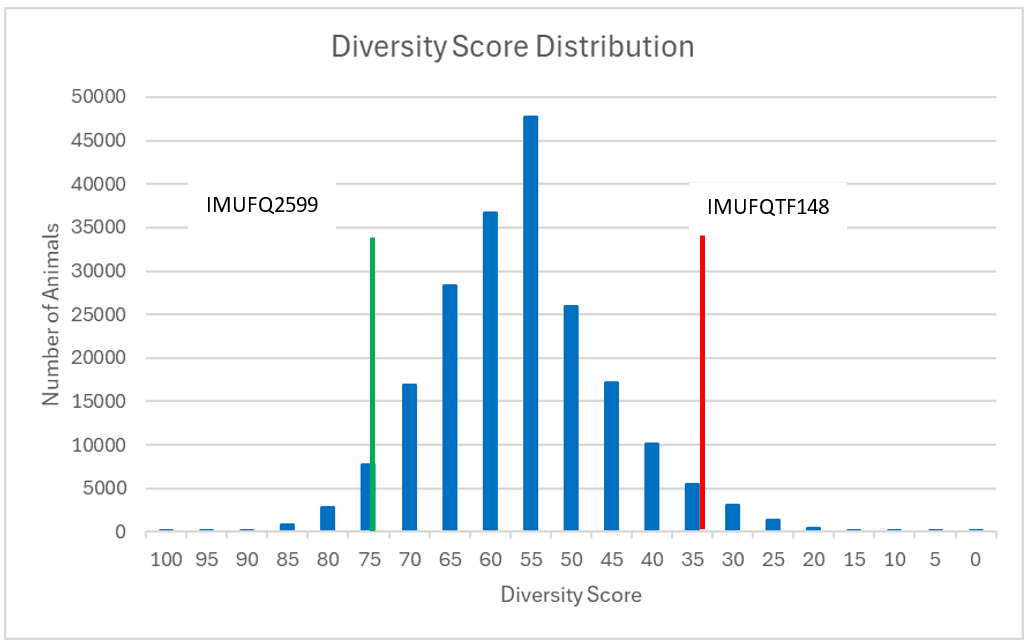
The Diversity Score Distribution graph shows that on average, the sire population has a Diversity Score average close to 50, but some animals have low diversity (eg. 25 or below) and are similar to the average of the current breeding female population. Some sires have high diversity (75 or above) and are genetically diverse from the average of the current female breeding population.
The foundation sire TF Yukiharunami 4 (IMUFQ2599) is highlighted in GREEN. He has 997 total progeny registered with the AWA, but only 24 females registered with the AWA within the last 5 calendar years. His genetic diversity score is relatively high compared to the current female breeding population.
For comparison, the foundation sire Itoshigenami (TF148) is shown is highlighted in RED. He has 7,797 progeny registered with the AWA, with 184 females registered with the AWA in the last 5 calendar years. TF148 has also sired many second and third generation influential sires that have high numbers of progeny registered within the last 5 calendar years. His genetic diversity score is relatively low compared to the current female breeding population.
Implications: When selecting bull calves for future breeding, a breeder can incorporate the Genomic Diversity Score as a selection criterion and decide to keep some high diversity calves as bulls even though their EBVs may only be average. This will help to increase genetic diversity and contain inbreeding in the future.
A breeder may also rank sires based on EBVs for a particular trait and then sorting them from highest to lowest Genomic Diversity to make science-based decisions on selecting breeding animals which are genomically different from the current cohort of breeding animals for the given level of EBVs.
The ability to calculate Genomic Diversity for suggested matings during a Matesel analysis is currently under development. This will improve a breeder’s ability to identify mating which would produce progeny with high genomic diversity at desired trait performance levels.
Implementation: Genomic Diversity will be published for all Fullblood Japanese Black animals with genomic (100K) profiles held by the AWA. It is not currently possible to analyse Genomic Diversity in Purebred, Wagyu content or in Red Wagyu animals due to the lack of effective reference populations.
Breeders will be able to use Genomic Diversity to assist in managing inbreeding and to accurately select for animals that provide true genetic diversity to their breeding herds.
